The core MIST deidentification system is implemented as a task
within the MAT toolkit. You will not use this core deidentification
task directly; rather, you'll be using a further specialization of the
core deidentification task, e.g., the AMIA or HIPAA task. Each task is
a child task of the parent task class, named "Deidentification", and
the implementation of each task is a subclass of the
Deidentification.DeidTaskDescriptor Python class. You can see evidence
of both these parent-child relationships in the task.xml file for your
particular deidentification task.
Creating new deidentification tasks is very, very complicated, and
we don't have the resources to document the process. Please consult the
specific deidentification tasks you have in your distribution and use
them as models if you absolutely need to build your own task.
The structural annotations you'll use (see the discussion of
annotations here)
are
just
the default structural annotations. The content annotations,
which are the ones you'll add or correct, correspond to the PII
categories for your task: e.g., the HIPAA categories for medical record
deidentification. Refer to the documentation for your specific
deidentification task for more details.
As we describe here,
steps
are
the
basic
activities
that
you
undertake
in
a
document.
E.g.,
the
tokenize step typically
identifies the word boundaries in the text. The typical
deidentification task has the following steps:
Note that the tokenize step found in the sample MAT task is not used
explicitly in MIST.
The nominate step requires
a replacer, which is a
strategy for generating replacement phrases. We discuss replacers below.
The typical deidentification task arranges these steps into a number
of workflows:
What's unusual about these four workflows is that the last of the
workflows applies to a different class of documents than the first
three. The resynthesize workflow is intended to apply to documents
which have been transformed into an obscure form, e.g., documents which
contain fillers like [PERSON] in place of the person names in the
original document. Under the resynthesize workflow, the tag step is a
simple pattern-matching operation, and the replacers convert obscured
documents into resynthesized documents. We discuss resynthesis below.
The steps of the deidentification task require the following step
implementations:
| Step implementation name |
Description |
|---|---|
| Deidentification.MultiZoneStepForUndo |
This step provides default undo
capabilities for the compound zone step described above. To be honest,
it's not exactly clear why this implementation is necessary. |
| Deidentification.ResynthZoneStep |
This step is a step which does
nothing, specifically in the case of zoning in the Resynthesize
workflow; no zoning is necessary, and we can't permit the default
zoning to apply. At the moment, the Resynthesize workflow does not
contain a zone step; this implementation may exist for historical
reasons. |
| Deidentification.ResynthTagStep |
The implementation of the tag
step in the Resynthesize workflow. This tag set merely needs to do
pattern-matching, rather than invoke an engine like Carafe. |
| Deidentification.NominateStep |
The implementation of the
nominate step. |
| Deidentification.TransformStep |
The implementation of the
transform step. |
As described here,
steps
can
also
take
key-value
pair
arguments
which
can
be
specified
in
the
task.xml
file or in the invocation of the MAT engine.
We document the arguments for these steps here.
The following key-value pairs might be generally useful for the
nominate step:
| Key |
Value |
Description |
|---|---|---|
| replacer |
one of the replacer UI names
shown below |
The name of the replacement
strategy to be used. If the workflow has more than one replacer
available, this setting is obligatory. This is most likely the only one
of these settings you'll ever use. |
| replacement_map |
a string |
You can customize your
replacement using a JSON string which describes a set of if-then rules
which you can apply to your clear -> clear match. See Customizing
deidentification replacement below. |
| replacement_map_file |
a filename |
You can also pass in your
replacement customization inside a file. |
The nominate step also accepts the following key-value pairs, but
they're pretty exotic and you're very unlikely to use them (they're
also not particularly well tested or supported):
| Key |
Value |
Description |
|---|---|---|
| cache_case_sensitivity |
a semicolon-separated sequence
of tag names, e.g. 'PERSON;LOCATION' |
In some cases, the replacer for
the given tag maintains an internal cache, to ensure, e.g., that
variations in names are replaced consistently throughout a document.
Names and institutions are the most obvious replacers which use a
cache. By default, the caches are not case-sensitive; this setting
allows the user to specify that some of them are. You will likely never
need this setting. |
| cache_scope |
a semicolon-separated sequence
of <tag>,doc|batch|none, e.g. 'PERSON,batch;LOCATION;doc' |
By default, if there is a cache
for a tag replacer strategy, its scope is the document; at each
document boundary, the cache is flushed. If you want to change this
scope, you can declare that it persists for the entire document batch
('batch') or turn off cacheing entirely ('none'). You will likely never
need this setting. |
| resource_file_repl |
a semicolon-separated sequence
of <file>=<repl> |
The replacement strategies are
driven by a large number of data files which are used as sources for
randomly created fillers. In some cases, you may want to replace these
files with files of your own. We're not going to document the way this
works, or how to use it, because it's just too arcane; the source file
core/python/ReplacementEngine.py in the deidentification source code
will help you understand how to use it. |
The transform step allows you to insert a prologue into your file.
| Key |
Value |
Description |
|---|---|---|
| prologue |
a string |
Specify the text of a prologue
to insert into the transformed document. You may wish to do this, e.g.,
to assert that all names in the document are fake. This option takes
preference over --prologue_file. |
| prologue_file |
a filename |
Specify a file which contains
the text of a prologue to insert into the transformed document. You may
wish to do this, e.g., to assert that all names in the document are
fake. The file is assumed to be in UTF-8 encoding. --prologue takes
preference over this option. If the filename is not an absolute
filename, it will be interpreted relative to the directory of the task
which is being trained for. (This is because this option more likely to
be provided in your task.xml file rather than on the command line.) |
Here's a standard use of MATEngine in this task. Let's say you want
to prepare a deidentified copy of a document with fake but realistic
English replacement PII elements, in rich format, and you have a
prologue in prologue.txt that you want to insert, and you have a
default model in your task. Here's how it works:
% $MAT_PKG_HOME/bin/MATEngine --task 'My Deid Task' --workflow Demo \
--steps 'zone,tag,nominate,transform' --replacer 'clear -> clear' \
--input_file /path/to/my/file.txt --input_file_type raw \
--output_file /path/to/my/resynth/file.txt.json --output_file_type mat-json \
--tagger_local --prologue_file prologue.txt
The --tagger_local option is required because by default, the
Carafe tagging task attempts to contact the tagging server.
The sample experiment described here
is a good place to start for constructing experiments for the
MATExperimentEngine. The primary differences you should keep in mind
are:
So if your task is named 'My Deid Task', then your simple experiment
might look like this:
<experiment task='My Deid Task'>
<corpora dir="corpora">
<partition name="train" fraction=".8"/>
<partition name="test" fraction=".2"/>
<corpus name="test">
<pattern>*.json</pattern>
</corpus>
</corpora>
<model_sets dir="model_sets">
<model_set name="test">
<training_corpus corpus="test" partition="train"/>
</model_set>
</model_sets>
<runs dir="runs">
<run_settings>
<args steps="zone,tag" workflow="Demo"/>
</run_settings>
<run name="test" model="test">
<test_corpus corpus="test" partition="test"/>
</run>
</runs>
</experiment>
In MIST 1.2, we've added a (very experimental) capability to
customize the deidentification replacement. This capability is
available only with clear -> clear replacement. To understand how to
use it, you'll need to know a bit more about how the replacement works.
Each replacement strategy consists of a digester and a renderer. The
digester produces a pattern which describes the features of the
digested element (e.g., for a phone number, was an area code present).
The digested pattern also contains the raw source filler, and, in the
case of the clear digester, the parsed form of the input in some cases
(e.g., for names and locations). The renderer uses the pattern to
generate its replacement, and, in the case of the clear renderer,
attempts to apply any customization rules it finds.
The customization rules are provided to the nominate step either
with the replacement_map or the replacement_map_file option. The
replacement map is a JSON string which has the format described
immediately below; the replacement_map_file provides a filename which
contains such a string.
The JSON string described a JSON hash (object) whose keys are the
file basenames which you're trying to deidentify; e.g., if you're
trying to deidentify /path/to/my/file.txt as in the example above, the
key for that file would be "file.txt". As a special case, if you're
accessing the MIST capability via the Web service, the name of the file
should be "<cgi>". The values of these keys should be another
JSON object whose keys are the names of the labels you're targeting. So
if you have rules for file.txt which target the DATE tag, your
replacement map will look like this so far:
{"file.txt": {"DATE": ...}}
The label key values are also JSON objects. These objects can have
two keys: caseSensitive (either true or false) and rules, which should
be a list of 2-element lists, where the first element of each sublist
is the antecedent and the second is the consequent. The antecedent and
consequent are each themselves JSON objects. The antecedent is a
recursive structure which describes a subset of the digested pattern;
the consequent contains two keys, seed and pattern, each of which
describe updates to the seed and pattern, respectively.
Much of the details of this needs to be derived from the source
code, but we provide a couple examples here.
Let's say you want to replace all occurrences of the last name
"Marshall" for the NAME tag in the file file.txt with the last name
"Bigelow". Your specification will look like this:
{"file.txt":
{"NAME":
{"rules": [[{"parse": {"lastName": "Marshall"}}, {"seed": {"lastName": "Bigelow"}}]]}}}
All eligible rules apply. So if you want to replace the first name
"Betty" with the first name "Phyllis", you can add another rule:
{"file.txt":
{"NAME":
{"rules": [[{"parse": {"lastName": "Marshall"}}, {"seed": {"lastName": "Bigelow"}}],
[{"parse": {"firstName": "Betty"}}, {"seed": {"firstNameAlts": ["Phyllis"]}}]]}}}
If you want to replace them only when they're together, do this:
{"file.txt":
{"NAME":
{"rules": [[{"parse": {"lastName": "Marshall", "firstName": "Betty"}},
{"seed": {"lastName": "Bigelow", "firstNameAlts": ["Phyllis"]}}]]}}}
If you only want to replace the literal string "Betty Marshall", do
this:
{"file.txt":
{"NAME":
{"rules": [[{"input": "Betty Marshall"},
{"seed": {"lastName": "Bigelow", "firstNameAlts": ["Phyllis"]}}]]}}}
(Note that the input key is available for all tags, while the
details of the parse and pattern keys differ from tag to tag).
Let's say you want to control how much dates are shifted (dates are
shifted by a consistent amount throughout a single document). Do this:
{"file.txt":
{"DATE":
{"rules": [[{}, {"pattern": {"deltaDay": 5}}]]}}}
The deltaDay attribute of the pattern controls the date shift, and
this rule applies the shift to all dates (because the antecedent is
empty).
Finally, let's say you want to do a consistent substitution of
certain IDs, e.g., patient IDs, for correlation with an external data
escrow application which manages deidentification for your structured
records. If your tag is IDNUM, you can do this:
{"file.txt":
{"IDNUM":
{"rules": [[{"input": "PATNO67897"}, {"seed": {"id": "ID9938273"}}]]}}}
In the future, we hope to flesh out and further document this
capability.
The MAT toolkit comes with a Java API which allows you to read and
write MAT JSON documents, and access services provided by the MATWeb application. You can
find documentation
for the Java API under the "Core developer documentation" in your
documentation sidebar.
One common use of the Java API is in incorporating deidentification
capabilities in a larger Java application. Here's a Java fragment which
shows you how to do that. Remember, you must make sure you have all the
jars in the following directories in your class path:
import java.util.*;
import org.mitre.mat.core.*;
import org.mitre.mat.engineclient.*;
String res = "";
/* Modify the URL as needed. Only the host and port are required. */
String url = "http://localhost:7801";
/* This should be the name of your task, as expted by MATEngine. */
String task = "HIPAA Deidentification";
/* This should be the name of the workflow, as expected by MATEngine. */
String workflow = "Demo";
/* This should be the step sequence to perform, as expected by MATEngine. */
String steps = "zone,tag,nominate,transform";
HashMap<String, String> attrMap = new HashMap<String, String>();
/* This should be the name of the replacer you want to use. */
attrMap.put("replacer", "clear -> clear");
MATDocument doc = new MATDocument();
/* Your input string is the argument to setSignal(). */
doc.setSignal("Hello World");
/* Here's where you connect to the server. */
MATCgiClient client = new MATCgiClient(url);
try {
MATDocument resultDoc = (MATDocument) client.doSteps(doc, task,
workflow, steps, attrMap);
res = resultDoc.getSignal();
} catch (MATEngineClientException ex) {
/* Handle the error. */
System.out.println("Processing failed: " + ex.getMessage());
}
/* Do something with the retrieved deidentified text. */
System.out.println(res);
As we describe here,
workspaces
are
actively-managed
directory
structures
which
encapsulate
the
standard
workflows
in
MAT.
Workspaces
in
the deidentification task
have two extra document folders:
In addition, the completed folder has one new operation, redact, which clears and populates
these two new folders in parallel, by applying a replacer to the
specified documents in the completed folder. This operation is intended
to apply only to original documents, to produce redacted documents; the
inverse resynthesize operation isn't available via the workspace.
The normal file and workspace mode document window configurations
described here. The
nomination
and transformation steps require enhancements to the UI, as shown in
this section. The document exhibited is drawn from the CMC free text
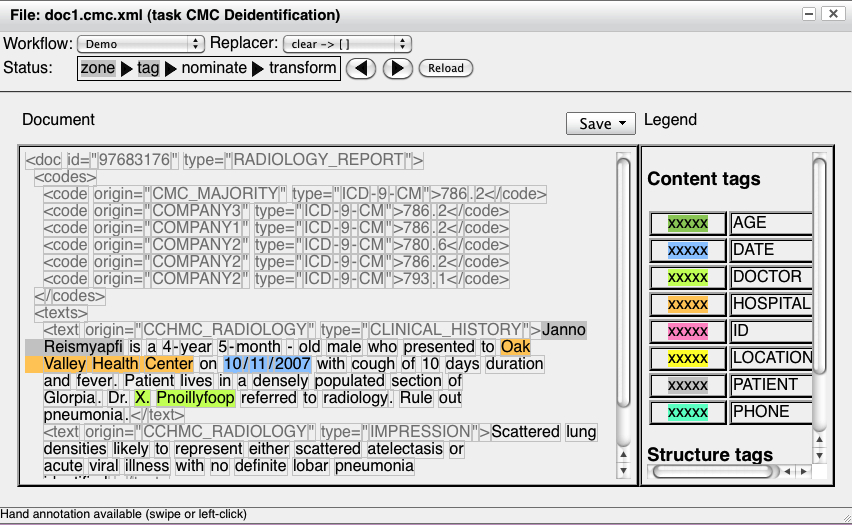
corpus of deidentified radiology reports.
First, we show the result of the tag step:

Note not just the additional steps in the workflow, but also the
menu labeled "Replacer" to the right of the workflow menu. This menu
allows you to select the replacement method for the PII elements. The
nominate step requires a replacer. Here's the result of the nominate
step:
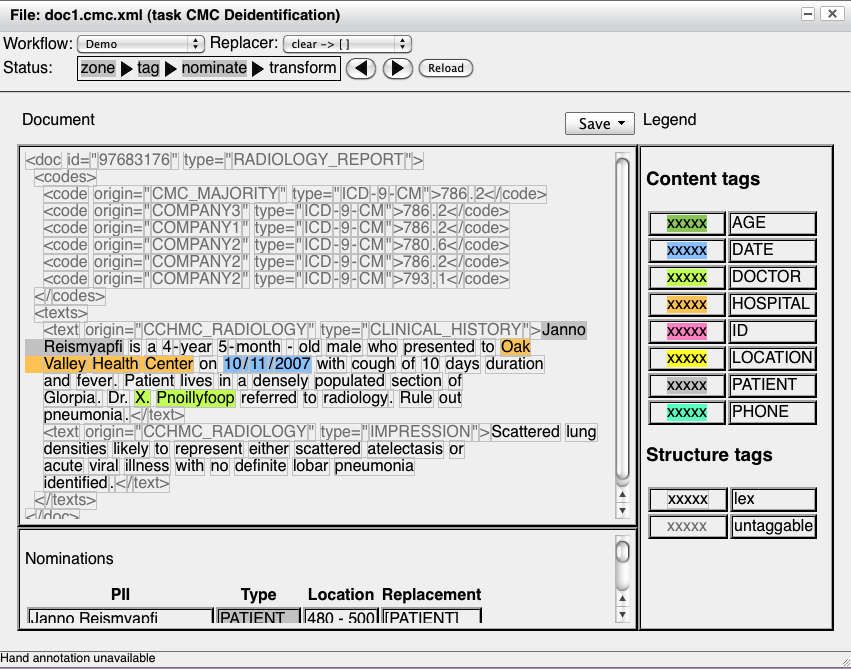
Note that the user has selected a replacer, and once the nominate
step is completed, a table appears below the document showing each PII,
along with its type, its location in the document, and its proposed
replacement. The final transform step inserts a second document pane:
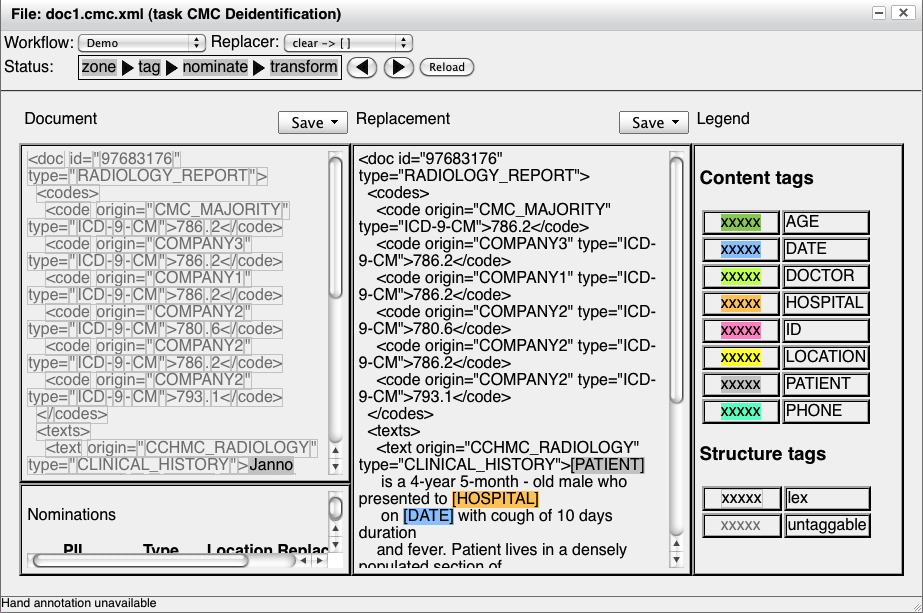
Note that the replacement document also has a menu to save the
replacement. These controls are
separate from the original document; i.e., if you select "mat-json"
from the "Save"
menu above the replacement document, the replacement document (not the
original document) will be saved as a rich MAT JSON document.
If you're in the workspace, the relevant point you'll notice a
difference is when viewing a document in the completed folder:

Like the file mode document window, this window has a replacer menu,
and the folder allows you to access the new redact operation. Unlike
file mode, the result of the redact operation will be a new document
window, rather than a new pane in the existing window.
The nomination step generates replacement fillers for the PII
elements identified by the content annotations. We identify two types
of replacement strategies:
The core deidentification system provides three basic types of
redaction:
The replacement engine first gathers a set of features from the input PII. For instance, in the case of names, it attempts to reproduce the number of tokens, the capitalization pattern, whether they correspond to a name with the last name first, etc. For dates, it attempts to preserve the offset from the earliest date in the document, as well as the specific details of how the date was formatted. Any features which cannot be determined from the input are assigned randomized values based on a weighted, hand-crafted estimation of the frequency of the possible values.
Once the feature values are determined, the engine generates replacement fillers. In the case of redaction, the fillers are trivially produced from the gathered features; in the case of resynthesis, the problem is considerably more complex, because our target is realistic English clear text. In the resynthesis case, the tokens for the replacement fillers are drawn from a variety of sources, including weighted lists of first and last names provided by the US Census, and lists of cities, states, streets, medical facilities and ZIP codes derived from various on-line resources. The replacement fillers are assembled based on the features the engine has already gathered; so, for instance, if the engine has determined that a name consists of a last name followed by a first name and an initial (as it might determine from a pattern like **NAME[AAA, BBB M] or a PII such as "Philips, Bruce R."), it will generate a new filler like “Ahmad, Jane Q”. Similarly, date offsets are preserved, so that the pair of dates Jan. 17 and Jan. 20 are shifted, but the 3 day difference is preserved. The engine also caches replaced name tokens on a document-by-document basis; so "AAA" will correspond to "Ahmad" throughout the document shown.
The resynthesis engine can be modified in a number of ways: where the replacement fillers are drawn from (e.g., an external lexicon or the corpus itself), how the fillers are constructed (whether they're replaced whole or reconstructed token-by-token), and how the fillers are cached for repetition (by document, by corpus, or without any cacheing). We will not document those customizations right now.
Each workflow in this task can be associated with replacers. This is
specified in the <settings> section of task.xml. The
specification pairs attribute-value settings, where the first
attribute-value pair is an arbitrary name of a replacer set and a
comma-separated list of replacer implementations, and the second
attribute-value pair is "<name>_workflows" and a comma-separated
list of workflow names. The interpretation is that the specified
replacers are available during the nomination step of the specified
workflows. Here's an example:
<settings>
<setting>
<name>redaction_replacers</name>
<value>BracketReplacementEngine.BracketReplacementEngine,CharacterReplacementEngine.CharacterReplacementEngine,ClearReplacementStrategy.ClearReplacementEngine</value>
</setting>
<setting>
<name>redaction_replacers_workflows</name>
<value>Demo,Hand annotation,Review/repair</value>
</setting>
<setting>
<name>resynthesis_replacers</name>
<value>BracketReplacementEngine.BracketResynthesisEngine</value>
</setting>
<setting>
<name>resynthesis_replacers_workflows</name>
<value>Resynthesize</value>
</setting>
</settings>
So the interpretation here is that the "Resynthesize" workflow has
the replacer BracketReplacementEngine.BracketResynthesisEngine
available, and the other three workflows have the other replacers
available.
Here are the available default replacers:
| Implementation |
UI name |
Description |
|---|---|---|
| BracketReplacementEngine.BracketReplacementEngine |
clear -> [ ] |
Maps clear text PIIs to the
bracketed name of the tag, e.g., "John Smith" -> "[PERSON]" |
| CharacterReplacementEngine.CharacterReplacementEngine |
clear -> char repl |
Obscures clear text PIIs by replacing alphanumeric characters with characters of the same type: "John Smith" -> "Pqty Muwqd" |
| ClearReplacementStrategy.ClearReplacementEngine |
clear -> clear |
Replaces clear text PIIs with
fake, synthesized but realistic clear text PIIs |
| BracketReplacementEngine.BracketResynthesisEngine |
[ ] -> clear |
Maps PIIs obscured with the
BracketReplacementEngine to clear text |
There are also replacers available to manage the De-ID-style
replacement, but those have to be customized in a child task.
At some point, you may want to define your own deidentification
task. We really don't recommend this, because much of the magic to make
this work is undocumented. If you must do this, start by copying the
entire directory structure of an existing deidentification task (your
distribution will contain at least one). Do not copy the core task.
Once you've copied the task directory, you'll need to make the
following changes to that task to so that MIST can recognize it: